Research
Cardiac Electrophysiology
Abnormal cardiac conduction or disrupted calcium homeostasis underlie/are associated with many cardiac disorders including cardiac arrhythmias, atrial fibrillation, and cardiac conduction disease (CCD). As the electrophysiological properties of the human heart are remarkably well conserved in teleosts, zebrafish embryos offer unique advantages to study the pathophysiological electrophysiological defects across the myocardial syncytium in a variety of mutants, each mimicking pathogenic/causal alleles/variants relevant to human cardiac disease.
Using custom-built microscopy which combines high resolution with unprecedented frequency, we are able to capture the electrical currents and calcium cycling within zebrafish hearts. We have successfully used these state-of-the-art techniques, combined with advanced immunohistochemistry (IHC) and secondary (electro)physiological assays in both adult zebrafish and larvae to pinpoint the exact biomechanical abnormalities in zebrafish deficient for genes associated with AFib, CCD, or Brugada syndrome.
Other efforts in the MacRae lab are focused on the impact of endocardial-to-myocardial signaling, aging, and metabolic disorders on cardiac conduction and calcium homeostasis throughout development.
Selected publications
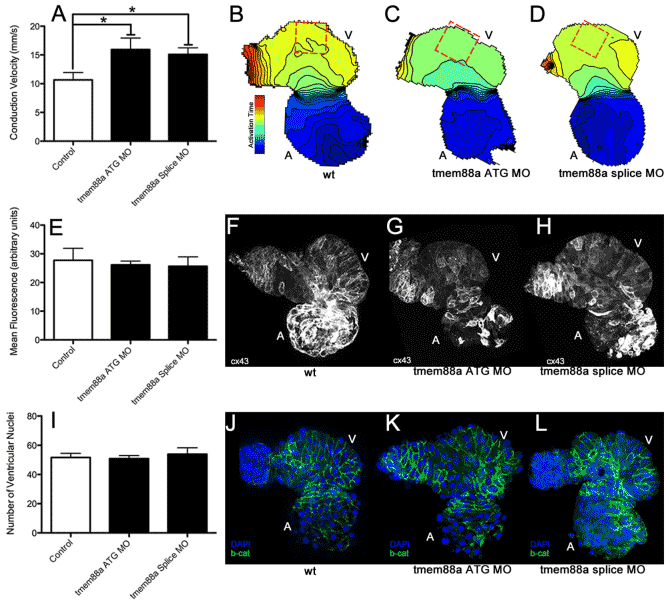
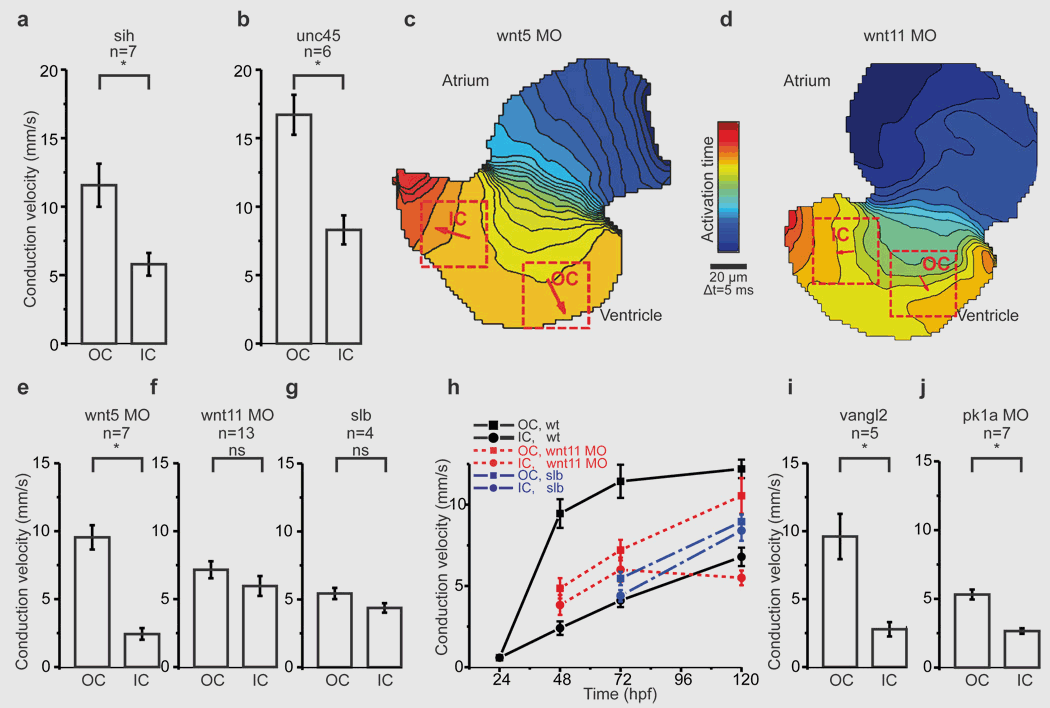
- Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca(2+) channel. Nature, 2011. View
- Metastable Atrial State Underlies the Primary Genetic Substrate for MYL4 Mutation-Associated Atrial Fibrillation. Circulation, 2020. View
- Noch1b and neuregulin are required for specification of the cardiac conduction system. Development, 2006. View
- LITAF (Lipopolysaccharide-Induced Tumor Necrosis Factor) Regulates Cardiac L-Type Calcium Channels by Modulating NEDD (Neural Precursor Cell Expressed Developmentally Downregulated Protein) 4-1 Ubiquitin Ligase. Circulation: Genomic and Precision Medicine, 2019. View
Cardiovascular Development and Disease
Cardiovascular development and physiology are dependent on a complex set of interactions between genetic and environmental factors, whose perturbation can result in early or late-onset cardiac and/or vascular pathologies. Our lab studies the etiology of cardiovascular disorders by looking at the kinetic and dynamic interplay between GWAS-derived loci/genes-of-interest and environmental cues associated with heart and vascular anomalies at both a system-wide, organ and single cell level. To do this, we utilize genetic and pharmacological approaches with subsequent high-resolution imaging, coupled to state-of-the-art techniques including (sc-)RNA and (sc-)ATAC sequencing, to gain further insights into the molecular pathways that drive cardiovascular pathologies.
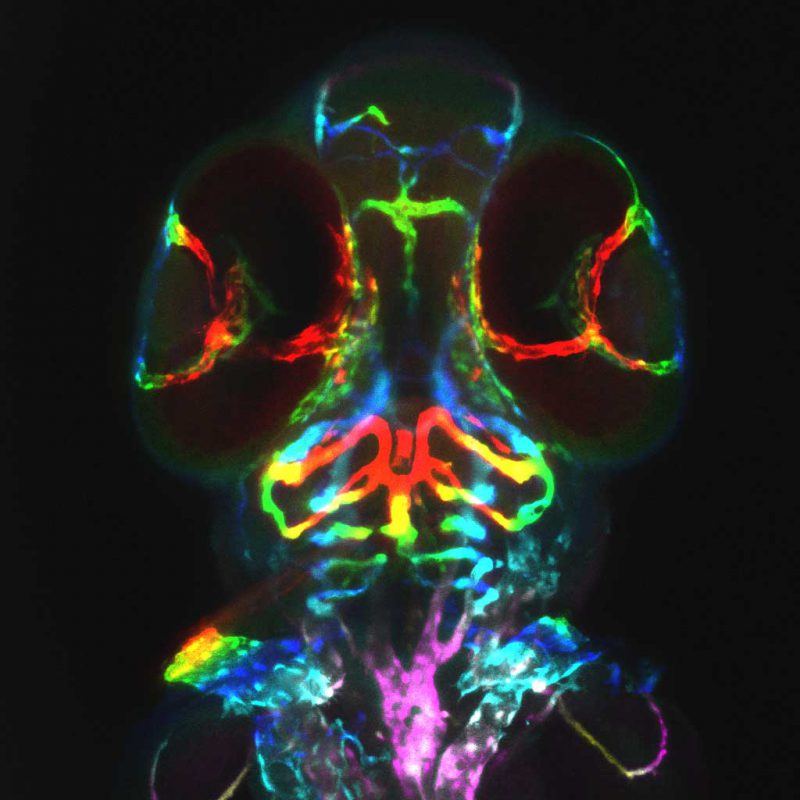
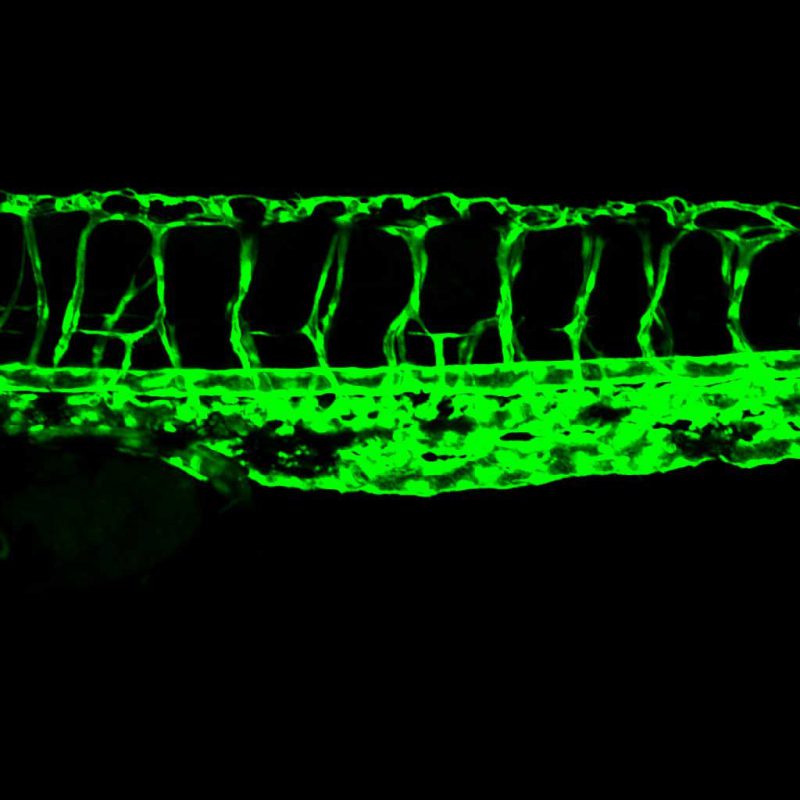
Phenotype-Genotype-Chemotype Predictions
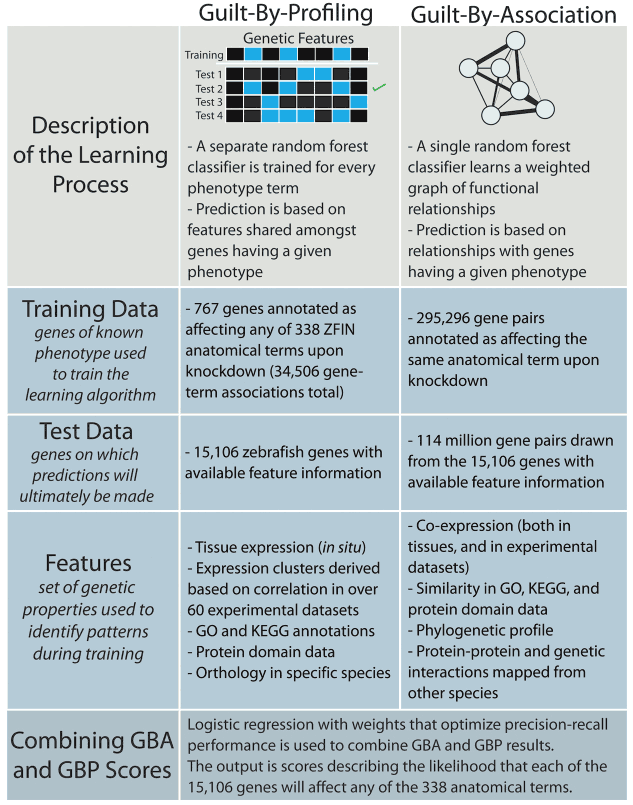
Both forward and reverse genetics in zebrafish have greatly advanced our understanding of gene function in various physiological and pathological settings. However, the vast number of surveyable genes and observable phenotypes require alternative strategies to maximize de novo discovery of gene functions. Through machine learning, we predicted the function of 15,000 zebrafish genes on 338 anatomical processes, with an emphasis on cardiovascular traits with high confidence. Ongoing efforts in the lab include the exploration of the biology behind these phenotype-genotype predictions and the expansion of this approach to predict phenotype-chemotype associations with the ultimate goal of enhancing drug discovery.
Selected publications
- Identification of specific metabolic pathways as druggable targets regulating the sensitivity to cyanide poisoning. PLoS One, 2018. View
- Generating and evaluating a ranked candidate gene list for potential vertebrate heart field regulators. Genom Data. 2015. View
- Novel cardiovascular gene functions revealed via systematic phenotype prediction in zebrafish. Development, 2014. View
- Selecting causal genes from genome-wide association studies via functionally-coherent subnetworks. Nat Methods, 2015. View
Drug Discovery Platform
Cardiovascular diseases (CVD) remain a main cause of death worldwide, but the heterogenous and complex nature of CVD has significantly hampered the development of novel, more efficacious therapies. Using defined zebrafish mutants that mimic causal alleles in patient cohorts, our lab has a long-standing interest in expediting drug discovery for diverse cardiovascular disorders. We use a myriad of in vivo approaches in zebrafish – ranging from simple biomarker assays to high-resolution imaging of complex cardiovascular traits and phenotypes – to screen thousands of small molecule compounds in a (semi-)automated fashion, with subsequent validation in mammalian systems. As such, we have identified compounds that prevent the onset of arrhythmogenic right ventricular cardiomyopathy, dilated cardiomyopathy, and atrial fibrillation among other CVD with high specificity and efficacy.
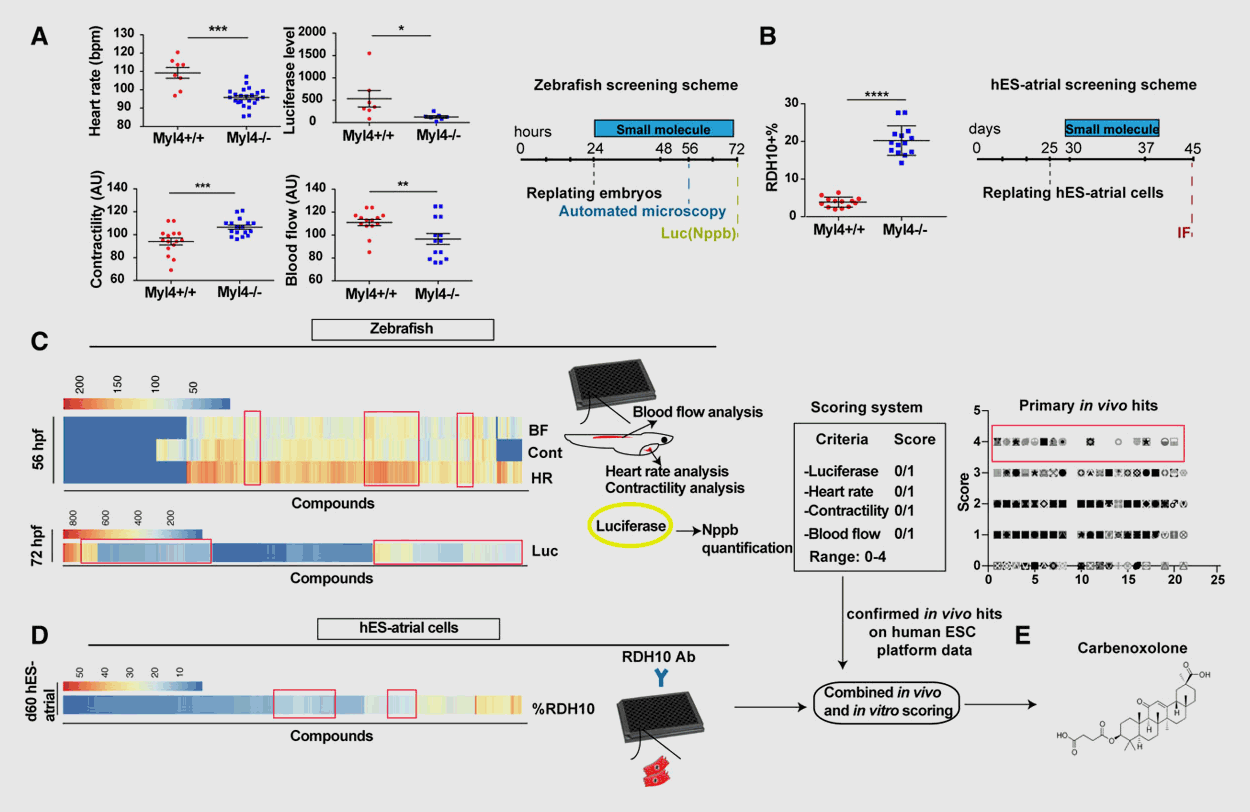
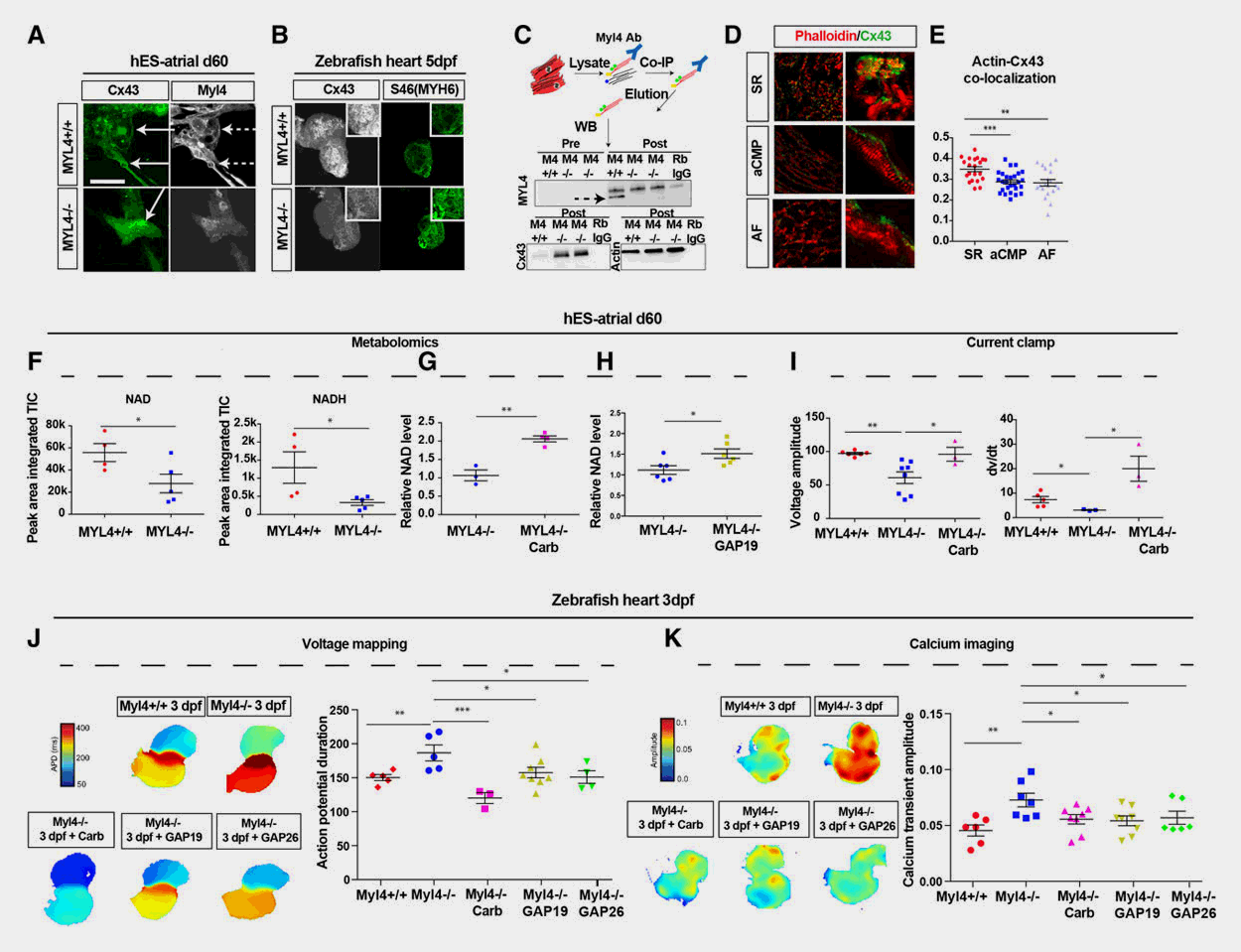
Selected publications
- Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Sci Transl Med., 2014. View
- Metastable Atrial State Underlies the Primary Genetic Substrate for MYL4 Mutation-Associated Atrial Fibrillation. Circulation, 2020. View
- In vivo natriuretic peptide reporter assay identifies chemical modifiers of hypertrophic cardiomyopathy signalling. Cardiovasc Res., 2011. View
- Using Zebrafish for High-Throughput Screening of Novel Cardiovascular Drugs. JACC Basic Transl Sci. 2017. View
Thrombosis
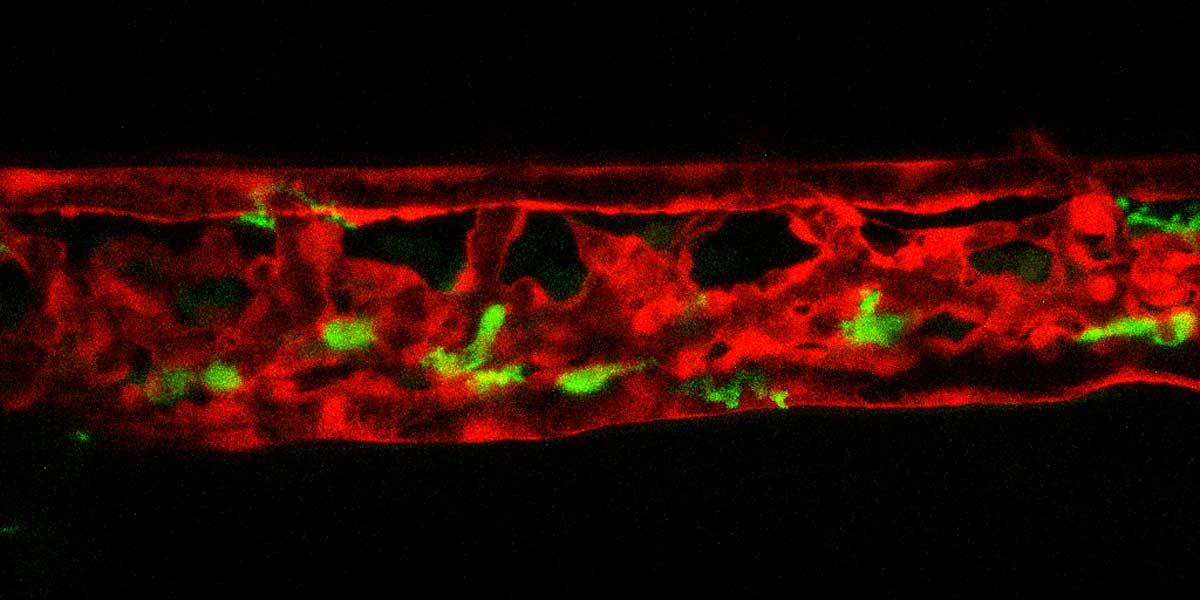
Cardiovascular defects and/or cardiometabolic diseases can lead to abnormal blood flow and increased shear stress on both hematopoietic and endothelial cells which can result in blood clotting (thrombosis). Although thrombosis is a major complication of metabolic and cardiovascular disease and remains a leading cause of death worldwide, our knowledge on the molecular and mechanical mechanisms that initiate clot formation remains limited. The MacRae lab has recently embarked on a project that aims to discover novel mechanical triggers in the clotting process, utilizing diverse assays to study pathogenic thrombosis in ex vivo human samples and defined zebrafish mutants at scale. Understanding force-driven mechanisms of thrombosis will allow us to isolate disease-linked prothrombotic triggers from general coagulation pathways, enabling the development of urgently needed precision therapies.
Identifying the Earliest Hallmarks of Atherogenesis
Although atherosclerosis commonly manifests itself in elderly patients, plaque formation starts decades earlier. However, most basic and clinical research focuses on slowing down the progression of plaque formation, rather than addressing the core molecular mechanisms at play that initiate this disease. Moreover, our knowledge on other phenotypes that might be more predictive for disease or unravel additional insights into disease etiology, remains limited.
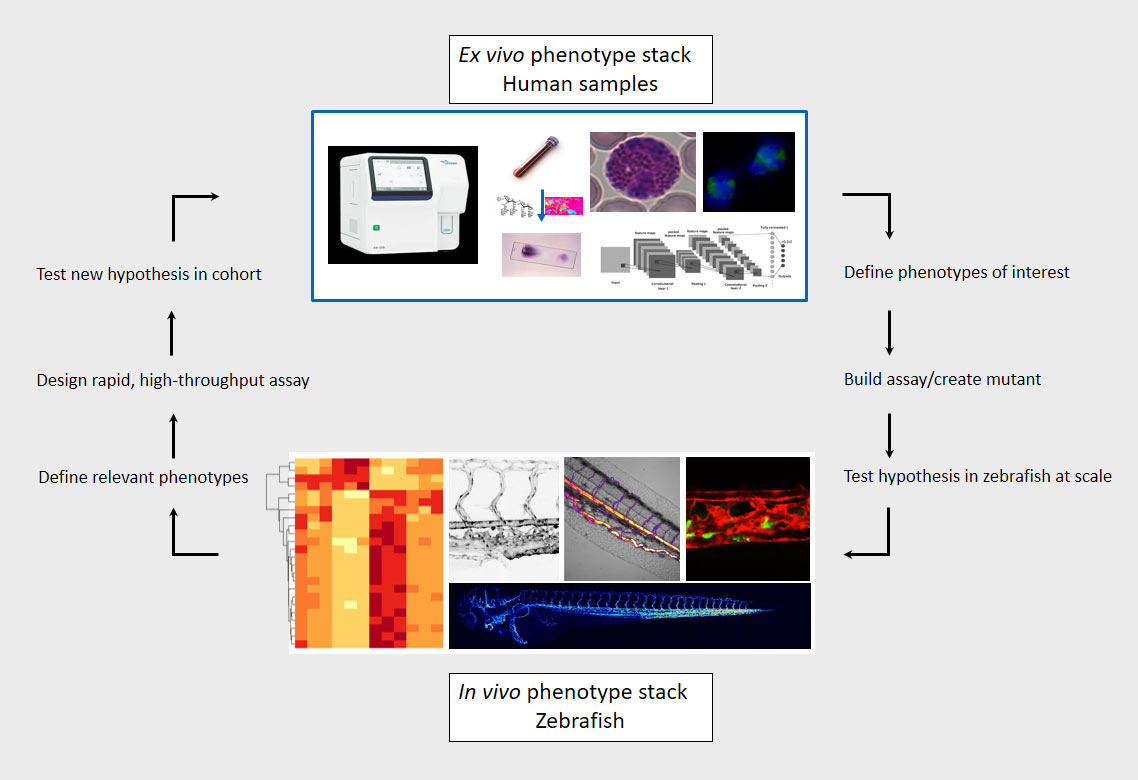
Our lab has built an array of zebrafish mutants that mimic some of the earliest clinical manifestations seen in familial hypercholesterolemia (FH) patients. We are currently studying the dynamic morphological, physiological, and transcriptional changes that occur in the diverse vascular, mural, and immunological cell types implicated in atherogenesis and their impact on disease progression in these mutants. Our lab employs a broad array of techniques to do this, including biochemical assays, (sc-)RNA/ATAC sequencing, and high-resolution microscopy. Through our interactive collaboration with One Brave Idea, we are uniquely positioned to test our zebrafish findings in patient derived samples. Likewise, we have multiple projects in the lab that use high-throughput sequencing and phenotyping of human blood samples to find novel phenotypes and genetic variants linked to atherosclerosis, which are subsequently validated in vivo using the zebrafish model.